Research
Whole-cell Modeling Approach
The assembly of whole-cell models spanning atomic to cellular resolutions is one of the grand challenges of the 21st century, and will require the convergence of biology, chemistry, physics, mathematics, computer science, and the digital arts. These models will change the way scientists approach biological research and drug design. We aim to provide a repository for β-cell data, connect researchers from different backgrounds, provide new insights into β-cell biology, and discover new approaches for designing therapies. Because diabetes affect hundreds of millions of people worldwide and there is still a need for development of more effective treatments. This makes β-cell biology of utmost importance to understand and design better diabetes therapeutics.
A diverse set of approaches and data types that span across spatial and temporal scales will be necessary for generating whole-cell models. Within the last few decades, researchers and clinicians have produced a wealth of relevant data detailing how these cells synthesize, pack and release insulin and what kind of complications occur during both Type I and Type II diabetes. However now is the time to connect all the literature along with new experimental and computational data and techniques to better understand the cell holistically and design effective therapeutics.
The assembly of these models will require an understanding of the basic ultrastructural architecture of the cell, a census of cell components (transcripts, proteins, lipids, metabolites, etc.), the abundance of components and their dynamic localization, and high-resolution structural information for all components and assemblies. The approaches we are using to collect this information are Cryo-electron tomography, X-ray tomography, Genome architecture mapping, Integrative structure modeling, Protein structure determination, Omics, Computational systems biology, Molecular graphics, and Live cell imaging.
Some of our early questions are:
- What happens to cellular ultrastructure during glucose or GLP-1R stimulation?
- How different β-cell lines and human primary β-cells differ in insulin response?
- How are insulin vesicles packed and organized spatially and temporally?
- How does all of this differ in disease-like states compared to healthy states?
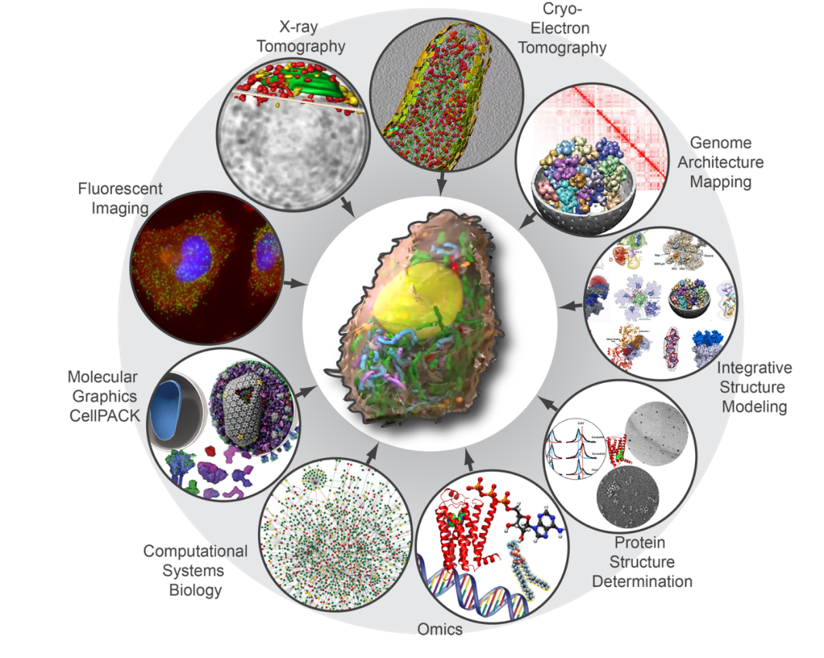
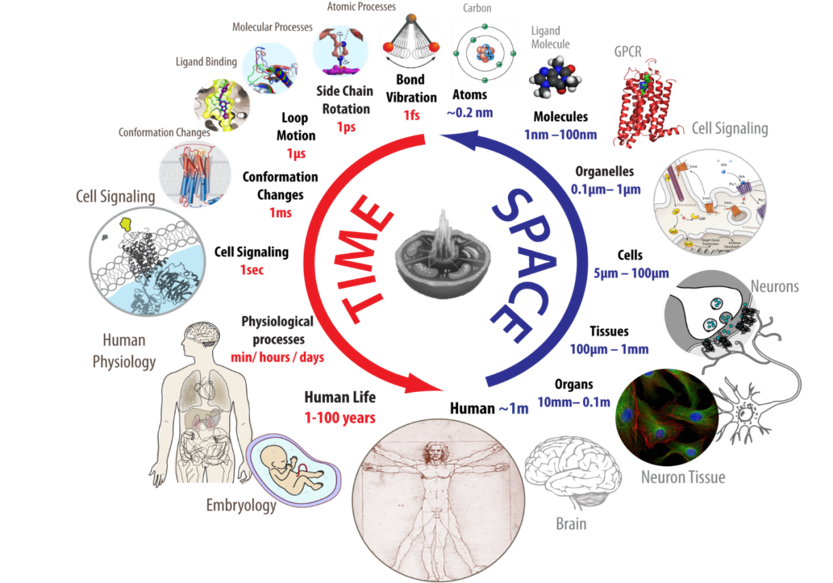
Opportunities and Challenges
- Community-wide open source effort to anyone interested in contributing data, models and/or ideas.
- Development of new tools and approaches to integrate data across scales and time.
- Merging experimental and computational data.
- Developing framework for assigning and measuring uncertainty/error/confidence for data/models in integrative approaches
World in a Cell
A broadly interdisciplinary team of scientists, storytellers, artists, programmers, and conceptual thinkers at the USC Bridge Institute at the Michelson Center for Convergent Biosciences and the USC School of Cinematic Arts are exploring the Pancreatic Beta Cell by creating a fully experiential virtual world in a single cell, using metaphors of the complex systems of a city. The goal is to use storytelling and world building to present scientific detail in ways that are engaging and approachable for both laymen and experts. By creating a virtual world inside a cell, based on the structure and function of a Pancreatic Beta Cell, we will allow people to explore a rich biochemical world while engaging concepts, pathways, and implications through narrative, all backed by scientific rigor.
Read More
Want to get involved?